A Simple Generator, Submitter and Organizer for Massive VASP Tasks
2024-11-20
Sometimes I need to submit VASP calculation tasks on different HPCs, and the environments of different nodes may vary. Some servers do not have batch submission scripts, so I took some time to write a script for personal use.
If you need to submit other types of computing tasks in bulk, it may also serve as a reference.
Principle Introduction
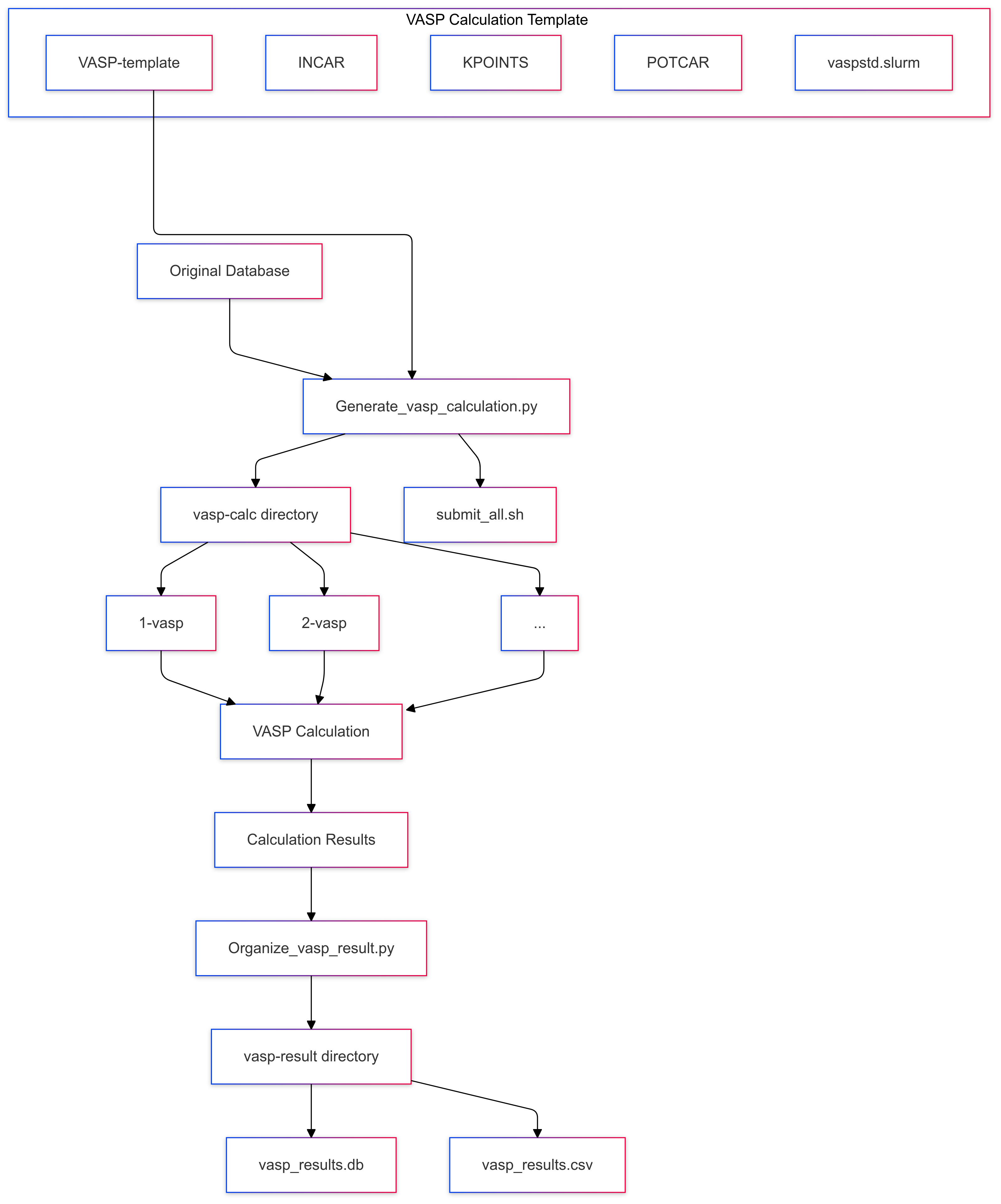
Overall Schematic Diagram
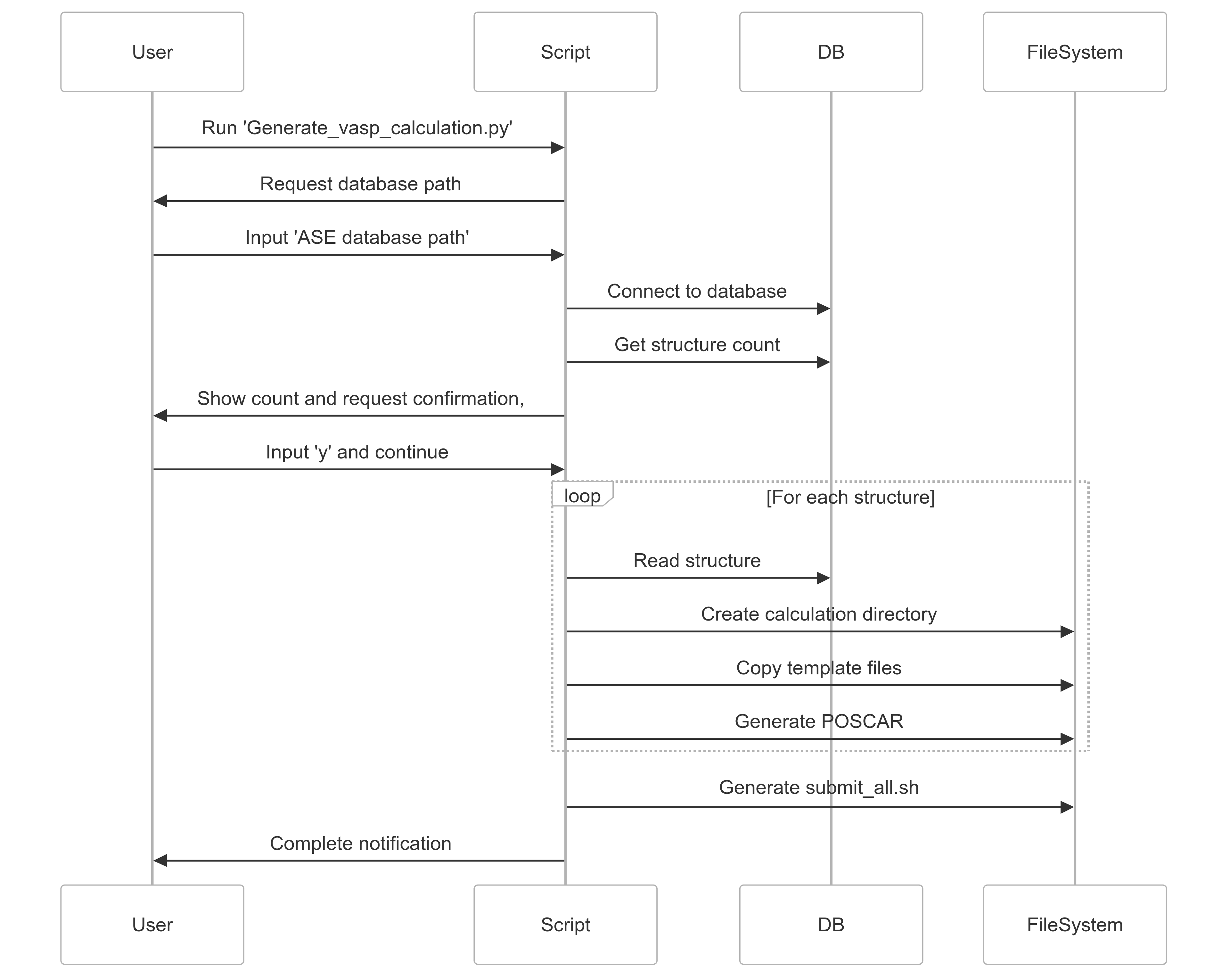
Generate_vasp_calculation.py
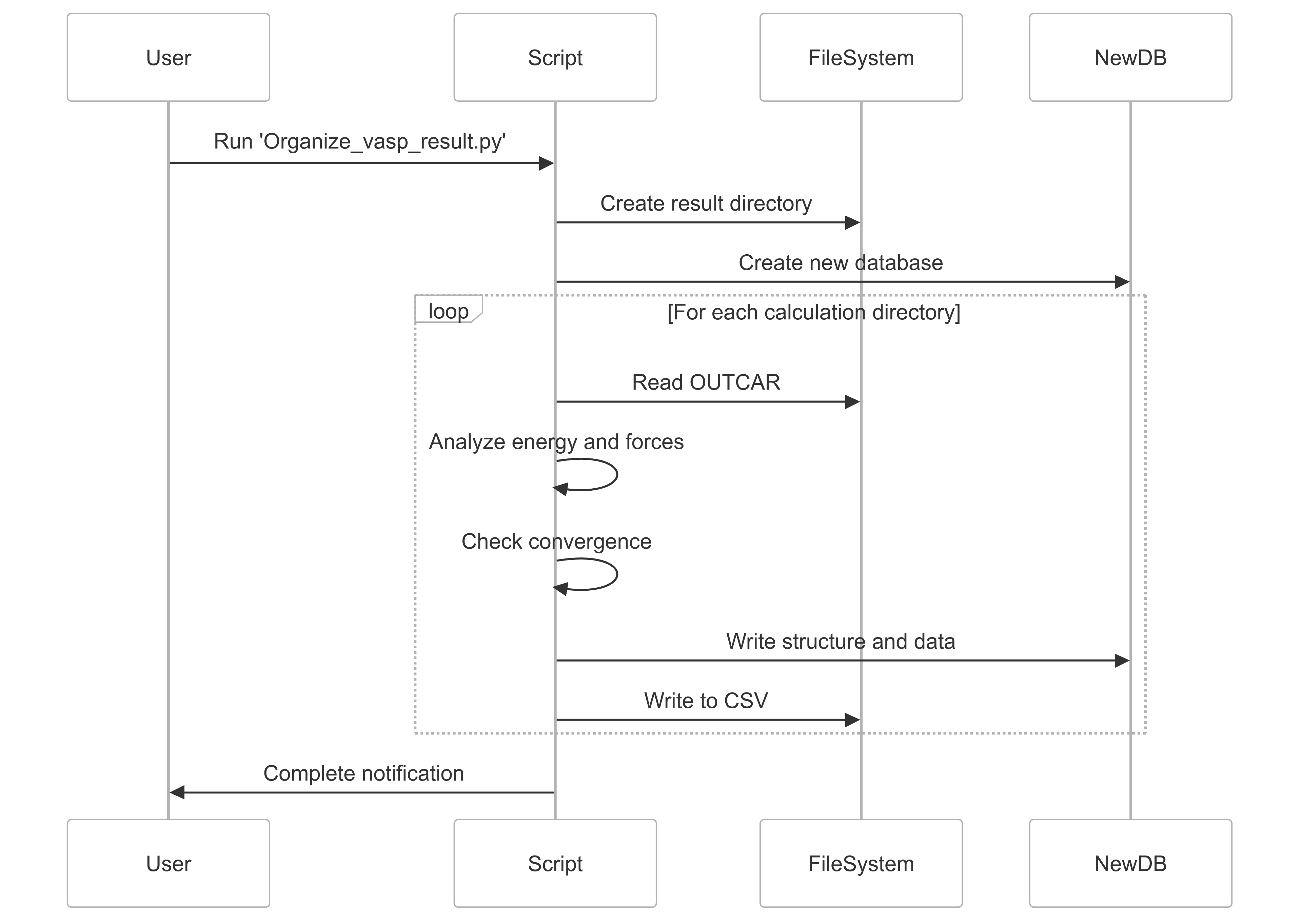
Organize_vasp_result.py
Usage Guide
Before Calculation: Generate Massive VASP Calculation Folder & Script for Submission
Generate_vasp_calculation.py
import os
import shutil
from ase.io import read, write
from ase.db import connect
def generate_vasp_calcs(db_path):
"""Generate VASP calculation folders and submission script"""
# Get absolute path of current working directory
work_dir = os.path.abspath(os.getcwd())
# Check if database file exists
if not os.path.exists(db_path):
raise FileNotFoundError(f"Database file not found: {db_path}")
# Check template directory
template_dir = "VASP-template"
required_files = ['INCAR', 'KPOINTS', 'POTCAR', 'vaspstd.slurm', 'vdw_kernel.bindat']
for file in required_files:
if not os.path.exists(os.path.join(template_dir, file)):
raise FileNotFoundError(f"Missing required file in template directory: {file}")
# Create vasp-calc directory
calc_base_dir = "vasp-calc"
if not os.path.exists(calc_base_dir):
os.makedirs(calc_base_dir)
# Connect to database
print(f"Reading database: {db_path}")
db = connect(db_path)
# Get total number of structures
n_structures = len(list(db.select()))
if n_structures == 0:
print("Warning: No structures found in database!")
return
print(f"\nFound {n_structures} structures in database")
confirm = input("Continue generating calculation files? (y/n): ")
if confirm.lower() != 'y':
print("Cancelled")
return
# Generate submission script
submit_lines = []
# Create calculation folder for each structure
count = 0
for row in db.select():
count += 1
# Create calculation directory
calc_dir = os.path.join(calc_base_dir, f"{row.id}-vasp")
if not os.path.exists(calc_dir):
os.makedirs(calc_dir)
# Copy template files
for file in required_files:
src = os.path.join(template_dir, file)
dst = os.path.join(calc_dir, file)
shutil.copy2(src, dst)
# Write structure to POSCAR
atoms = row.toatoms()
poscar_path = os.path.join(calc_dir, 'POSCAR')
write(poscar_path, atoms, format='vasp', direct=True, vasp5=True)
# Add submission command (using absolute path)
abs_calc_dir = os.path.join(work_dir, calc_dir)
submit_lines.append(f'cd "{abs_calc_dir}" && sbatch vaspstd.slurm')
print(f"Prepared calculation folder: {calc_dir}")
# Generate submission script (same level as vasp-calc)
submit_script = "submit_all.sh"
with open(submit_script, 'w') as f:
f.write("#!/bin/bash\n\n")
# Each line contains submission command and delay
for line in submit_lines:
f.write(f"{line} && sleep 0.3\n")
# Set script executable permission
os.chmod(submit_script, 0o755)
print(f"\nProcessed {count} structures")
print(f"Submission script generated: {submit_script}")
print("Run the following command to submit all calculations:")
print(f"bash {submit_script}")
if name == "__main__":
while True:
# Interactive input for database path
db_path = input("\nEnter database file path (q to quit): ")
if db_path.lower() == 'q':
print("Program exited")
break
# Check if file exists
if not os.path.exists(db_path):
print(f"Error: File '{db_path}' not found")
continue
# Check file extension
if not db_path.endswith('.db'):
print("Warning: File extension is not .db")
confirm = input("Continue anyway? (y/n): ")
if confirm.lower() != 'y':
continue
try:
generate_vasp_calcs(db_path)
break
except Exception as e:
print(f"Error: {str(e)}")
continue
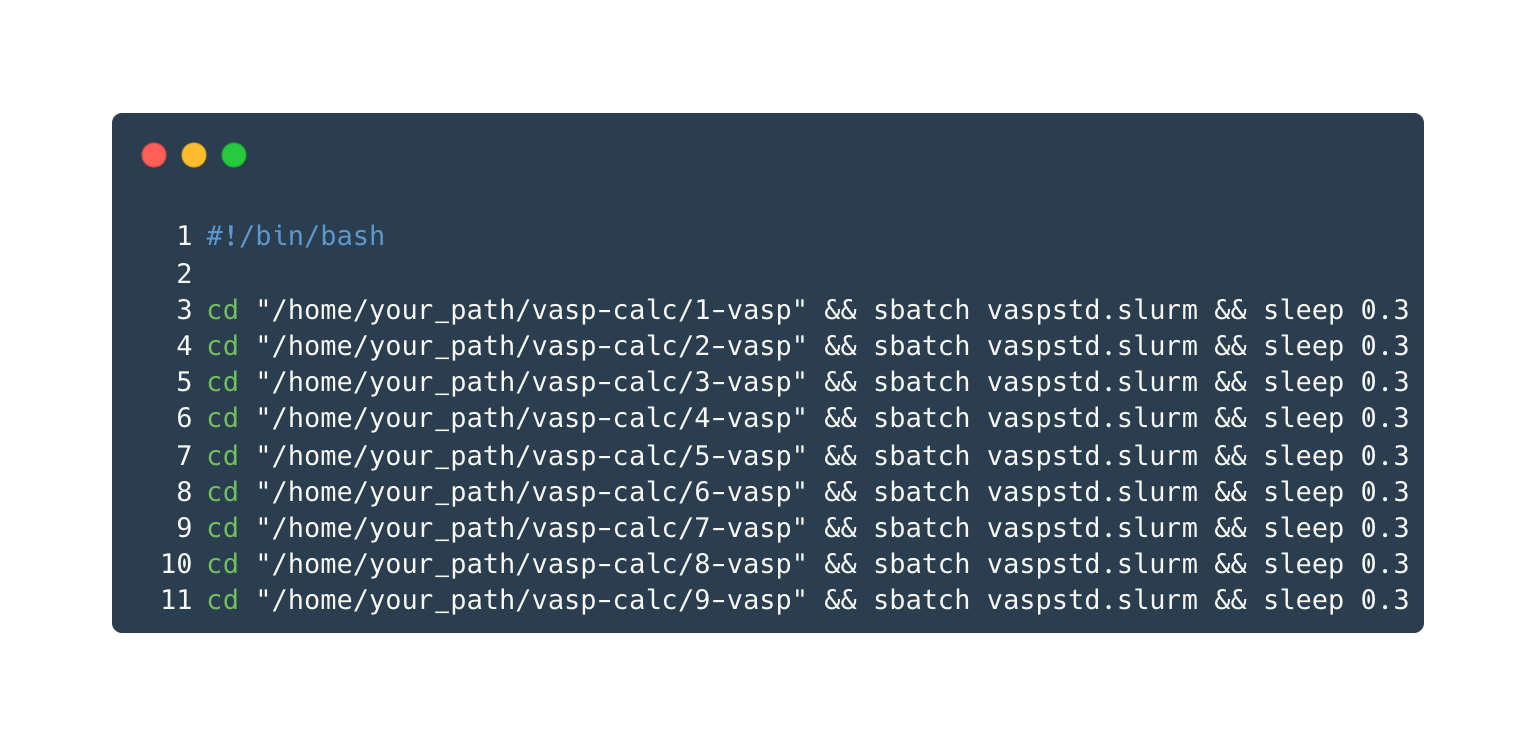
Submit Calculation Tasks
Just need to execute the script submit_all.sh
After Calculation: Organize VASP Result
Organize_vasp_result.py
from ase.io import read
from ase.db import connect
import os
import numpy as np
import pandas as pd
from tqdm import tqdm
from datetime import datetime
from pathlib import Path
from ase.io.vasp import read_vasp_out
import concurrent.futures
def get_path_similarity_key(path):
"""Sort paths based on numbers in directory names
Example: 'vasp-calc/1-vasp/OUTCAR' will be sorted by number 1
"""
try:
parts = Path(path).parts
for part in parts:
if '-vasp' in part:
number = int(part.split('-')[0])
return number
return float('inf')
except:
return float('inf')
def is_calculation_converged(outcar_path):
"""Check if VASP calculation has converged"""
try:
with open(outcar_path, 'r') as f:
f.seek(max(0, os.path.getsize(outcar_path) - 50000))
content = f.read()
return "reached required accuracy" in content
except:
return False
def read_last_frame(outcar_path):
"""Read only the last frame from OUTCAR"""
try:
atoms = read_vasp_out(outcar_path, index=-1)
return atoms
except Exception as e:
print(f"Error reading file {outcar_path}: {str(e)}")
return None
def process_last_frame(outcar_path):
"""Process the last frame of OUTCAR file"""
try:
last_frame = read_last_frame(outcar_path)
if last_frame is None:
return None
converged = is_calculation_converged(outcar_path)
energy = last_frame.get_potential_energy()
forces = last_frame.get_forces()
forces_sq = np.sum(forces**2, axis=1)
max_force = float(np.sqrt(forces_sq.max()))
n_atoms = len(last_frame)
data = {
'energy': float(energy),
'forces': forces.tolist(),
'forces_max': max_force,
'calculation_id': os.path.basename(outcar_path),
'step_number': -1,
'pbc': last_frame.pbc.tolist(),
'cell': last_frame.cell.array.tolist()
}
if last_frame.get_stress() is not None:
data['stress'] = last_frame.get_stress().tolist()
if last_frame.has('initial_magmoms'):
data['magmoms'] = last_frame.get_initial_magnetic_moments().tolist()
data['outcar_path'] = os.path.relpath(outcar_path)
data['converged'] = converged
data['n_atoms'] = n_atoms
cpu_time = get_cpu_time(outcar_path)
data['cpu_time'] = cpu_time
return data, last_frame
except Exception as e:
print(f"Error processing file {outcar_path}: {str(e)}")
return None
def get_cpu_time(outcar_path):
"""Get CPU time from OUTCAR file"""
try:
with open(outcar_path, 'r') as f:
f.seek(max(0, os.path.getsize(outcar_path) - 50000))
content = f.read()
for line in content.split('\n'):
if "Total CPU time used (sec):" in line:
return float(line.split(':')[1].strip())
except:
pass
return 0.0
def main():
start_time = datetime.now()
print(f"\nStart time: {start_time}")
output_dir = "./vasp-result"
os.makedirs(output_dir, exist_ok=True)
db_path = os.path.join(output_dir, "vasp_results.db")
csv_path = os.path.join(output_dir, "vasp_results.csv")
vasp_dirs = [
"./vasp-calc",
# "./other-floors-available",
]
print("Search directories:")
for dir_path in vasp_dirs:
print(f"- {os.path.abspath(dir_path)}")
print(f"Database path: {os.path.abspath(db_path)}")
print(f"CSV file path: {os.path.abspath(csv_path)}")
vasp_files = []
for vasp_dir in vasp_dirs:
if not os.path.exists(vasp_dir):
print(f"Warning: Directory {vasp_dir} does not exist, skipped")
continue
for root, dirs, files in os.walk(vasp_dir):
for file in files:
if file == 'OUTCAR':
vasp_files.append(os.path.join(root, file))
vasp_files.sort(key=get_path_similarity_key)
print(f"Found {len(vasp_files)} OUTCAR files")
total_count = 0
converged_count = 0
with open(csv_path, 'w') as f:
header = 'ID,outcar_path,energy,forces_max,n_atoms,converged,cpu_time\n'
f.write(header)
with connect(db_path, append=False) as db:
for i, outcar_path in enumerate(tqdm(vasp_files, desc="Processing OUTCAR files"), start=1):
result = process_last_frame(outcar_path)
if result:
data, atoms = result
data['ID'] = i
db.write(atoms, data=data)
csv_line = f"{i},{data['outcar_path']},"
csv_line += f"{data['energy']},{data['forces_max']},"
csv_line += f"{data['n_atoms']},{1 if data['converged'] else 0},"
csv_line += f"{data['cpu_time']}\n"
f.write(csv_line)
total_count += 1
if data['converged']:
converged_count += 1
if os.path.exists(csv_path):
file_size = os.path.getsize(csv_path)
print(f"\nCSV file created: {os.path.abspath(csv_path)}")
print(f"File size: {file_size} bytes")
print("\nCSV file preview:")
with open(csv_path, 'r') as f:
head = [next(f) for _ in range(5)]
print(''.join(head))
else:
print(f"\nWarning: CSV file not created: {csv_path}")
print("\nProcessing statistics:")
print(f"Successfully processed files: {total_count}")
print(f"Converged calculations: {converged_count}")
print(f"Non-converged calculations: {total_count - converged_count}")
end_time = datetime.now()
print(f"\nEnd time: {end_time}")
print(f"Total time: {end_time - start_time}")
with connect(db_path) as db:
print(f"\nTotal entries in database: {len(db)}")
if name == "__main__":
main()